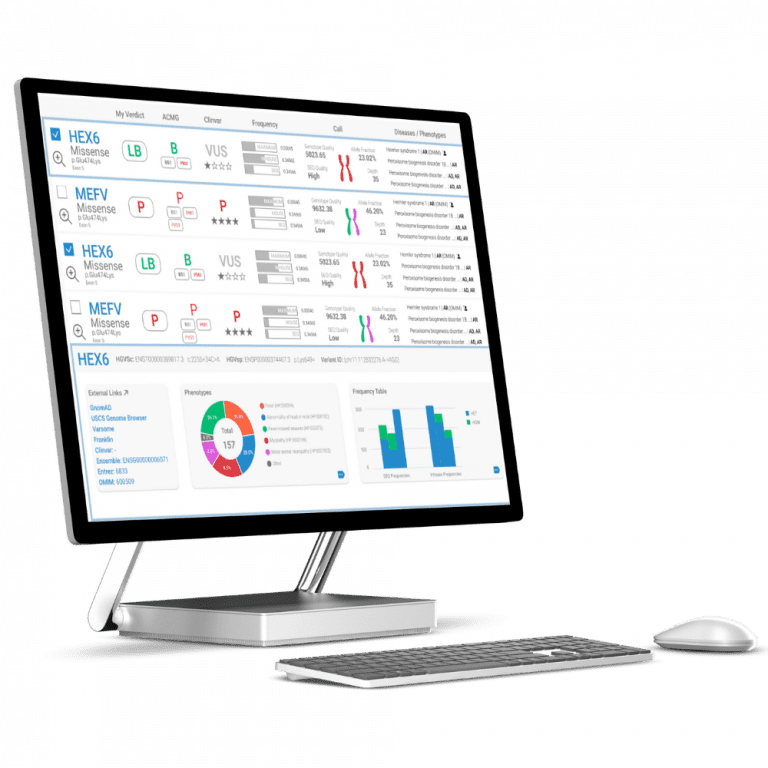
Turning genomic data into actionable insights
From raw data to diagnosis, SEQ Platform provides comprehensive NGS data analysis and accurate ACMG classification for confident variant interpretation.

Why choose us?
Increase your Diagnosis Rate
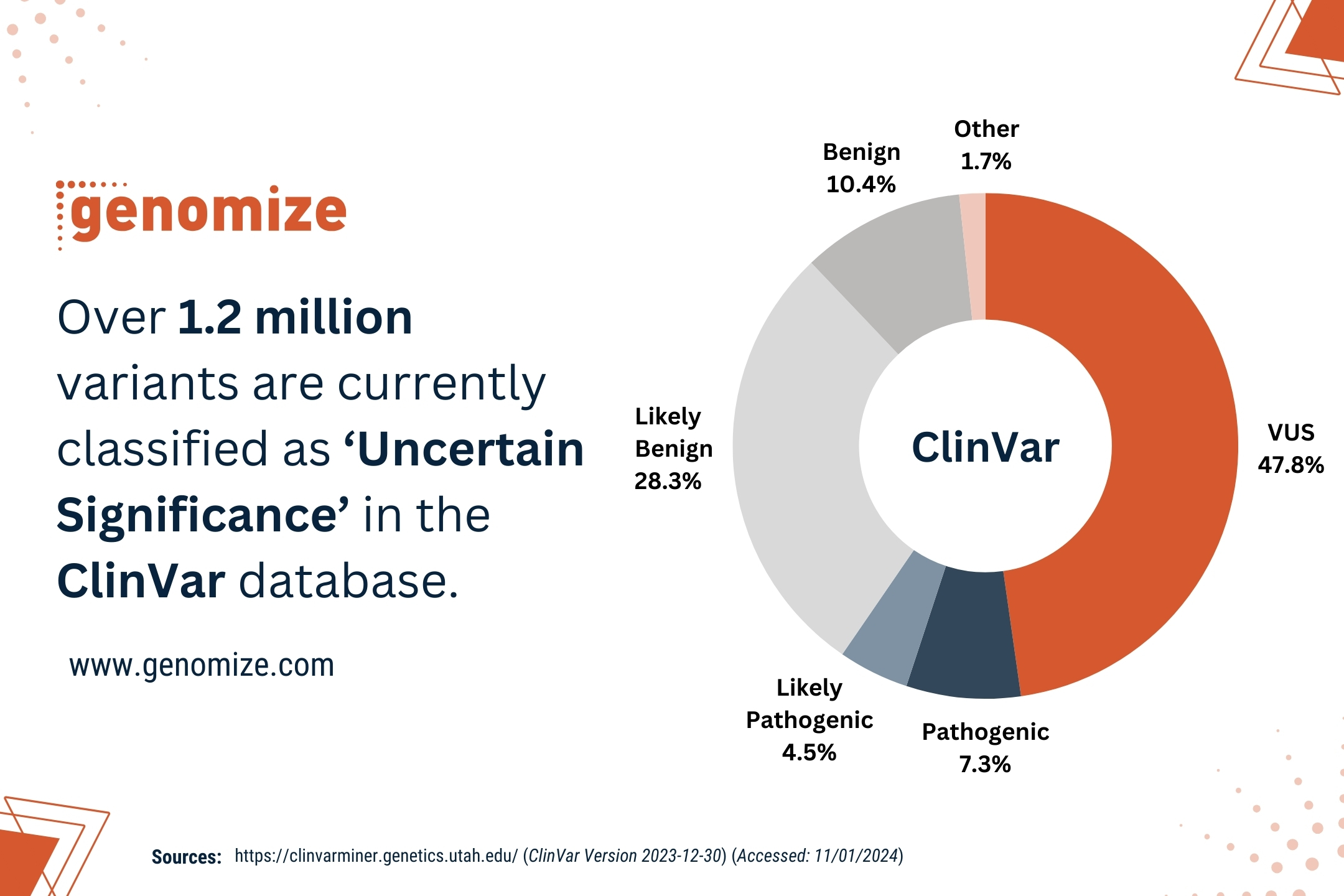
The SEQ Platform’s advanced bioinformatics pipelines and AI-driven variant prioritization enable accurate detection of genomic variants, leading to more precise diagnosis of genetic disorders.
Build your own Real-time Genotype/Phenotype Database
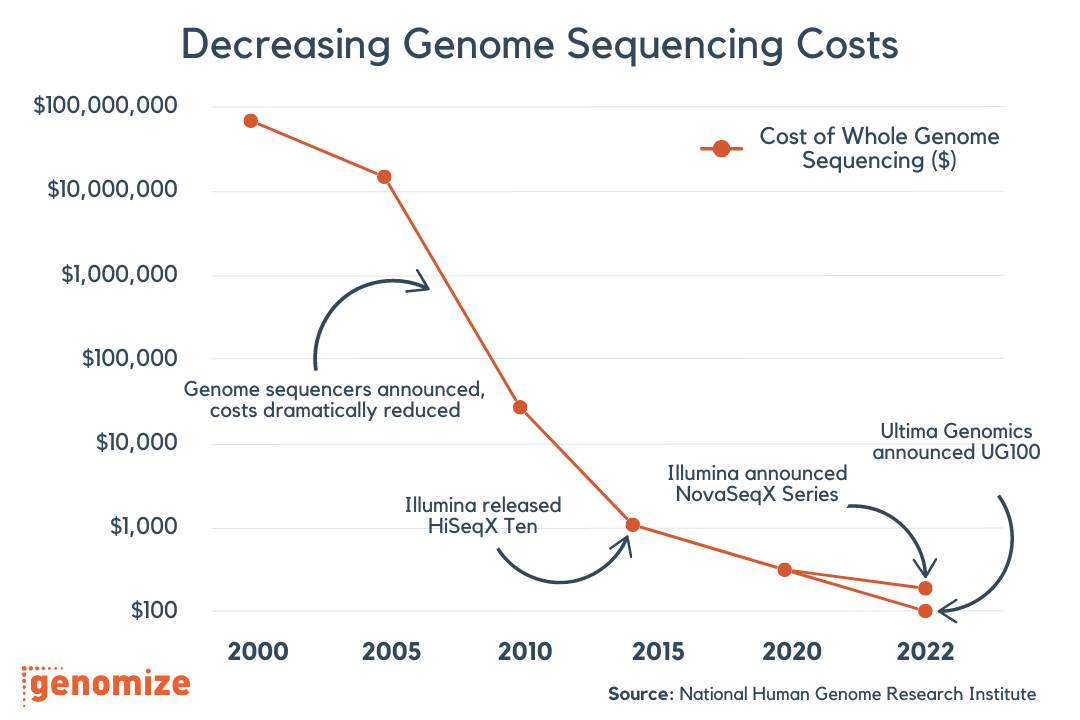
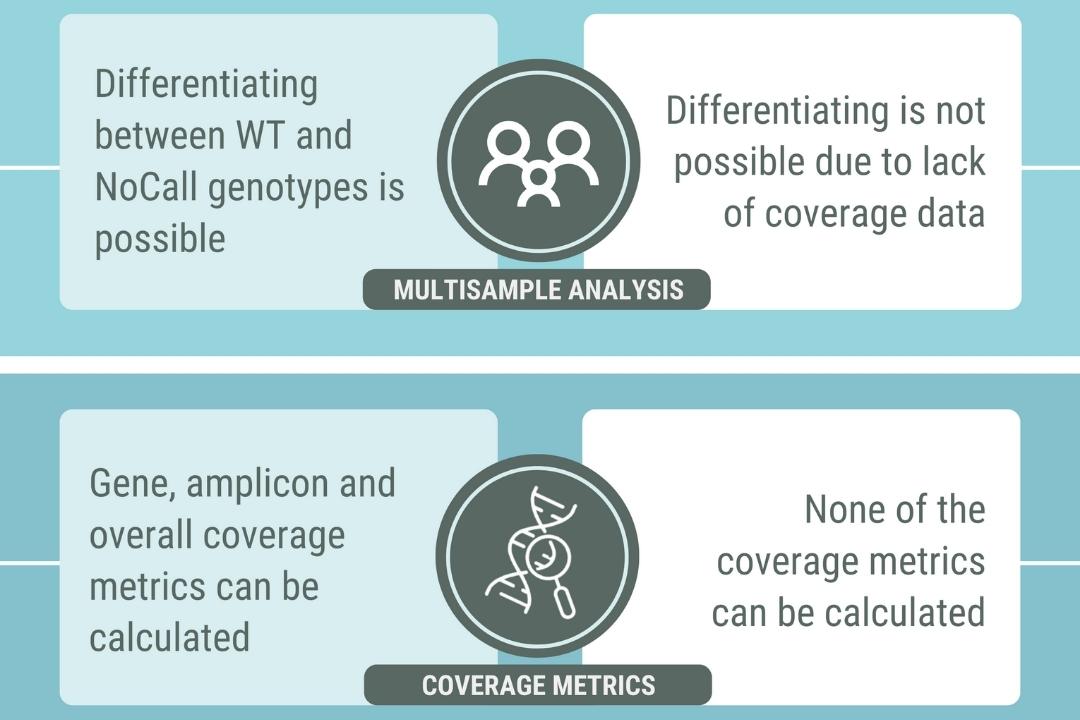
Take control of your genomic data. With the SEQ Platform, you can easily build and manage your own genotype/phenotype database, empowering you to make informed decisions based on your unique genomic insights.


Find actionable alterations and treatment options for cancer patients
SEQ Platform’s highly sensitive somatic pipeline can help identify alterations and provide insights into potential treatment options based on ASCO/AMP/CAP guidelines.