FastQ and VCF (Variant Call Format) are two different file formats used in bioinformatics analysis of the DNA sequencing data, but they serve different purposes and contain different types of information.
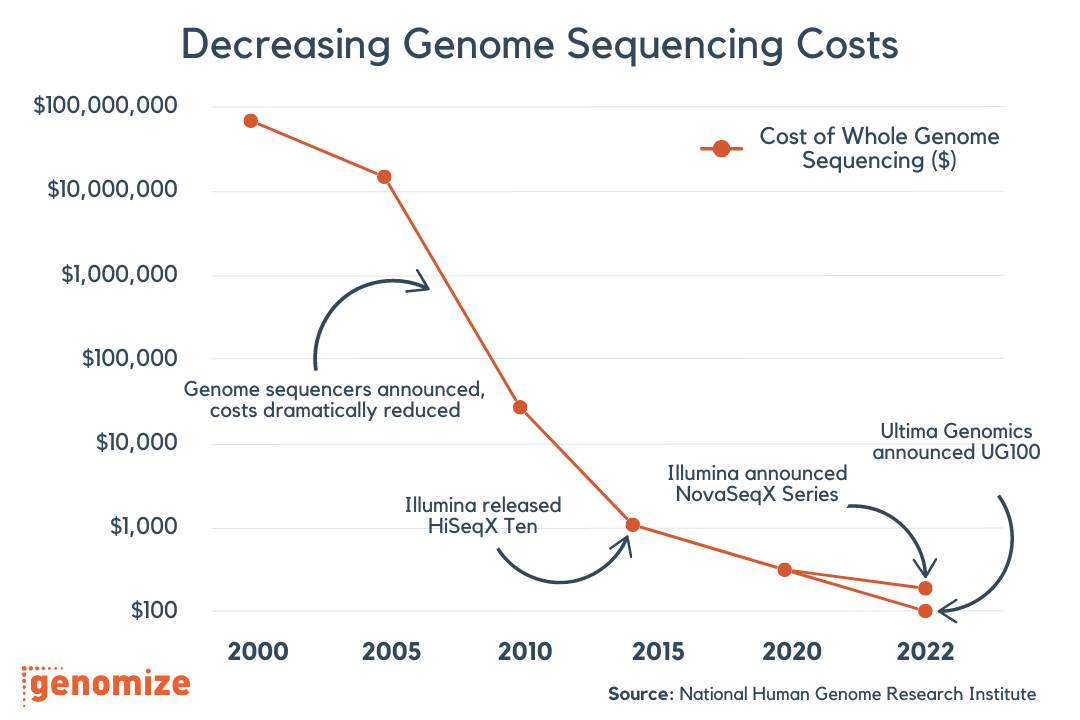
FastQ files are the raw output files generated by NGS platforms, such as Illumina, MGI, or Ion Torrent. These files store the sequence reads along with their corresponding quality scores. The quality scores provide an estimate of the confidence level for each base call, which is crucial for downstream analysis.
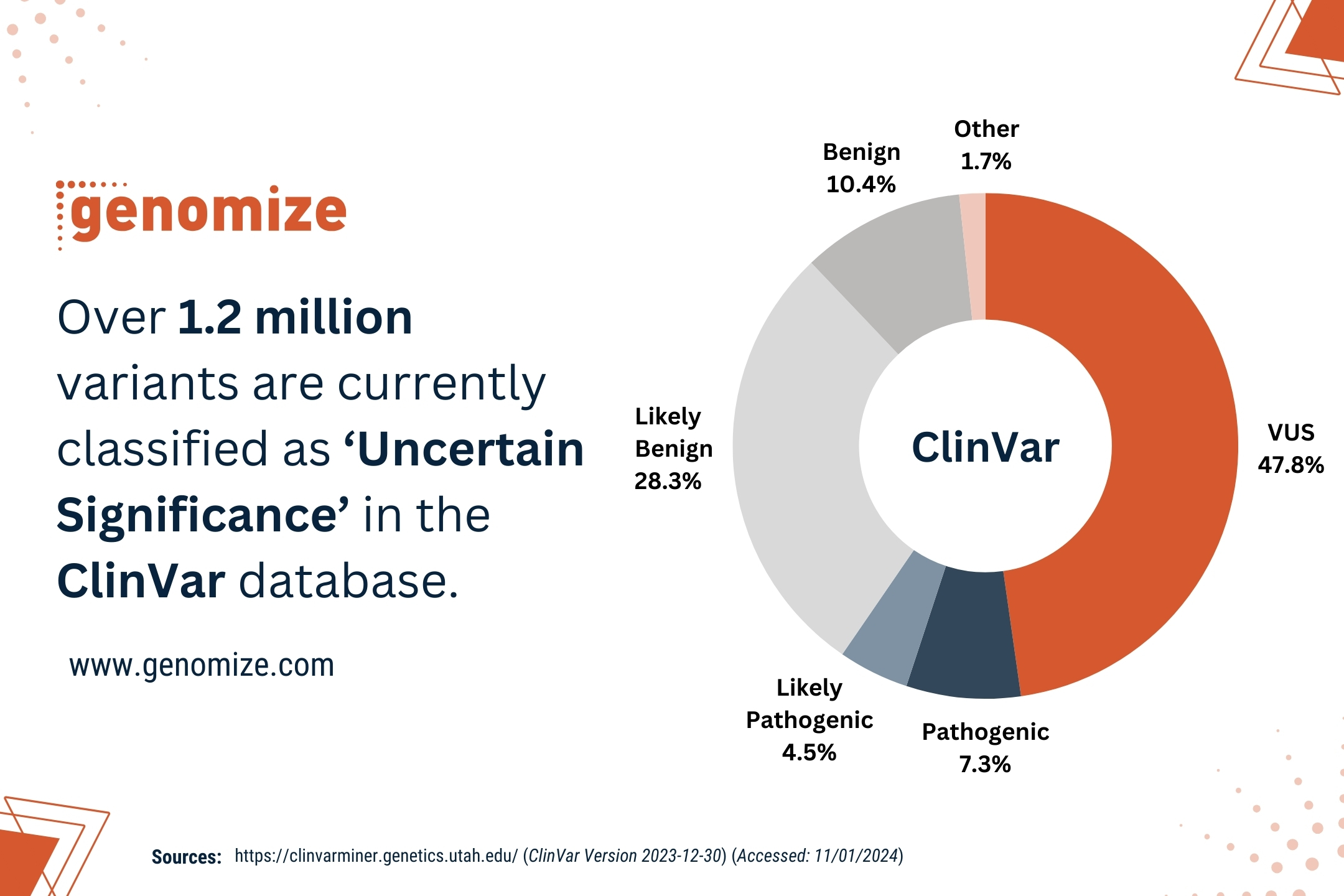
On the other hand, VCF files store information about genetic variants that have already been identified and called from the sequence data, meaning that VCF is a form of processed data. VCF files typically contain information about single nucleotide polymorphisms (SNPs), insertions, deletions, and other structural variants, along with their genomic coordinates and additional annotations.
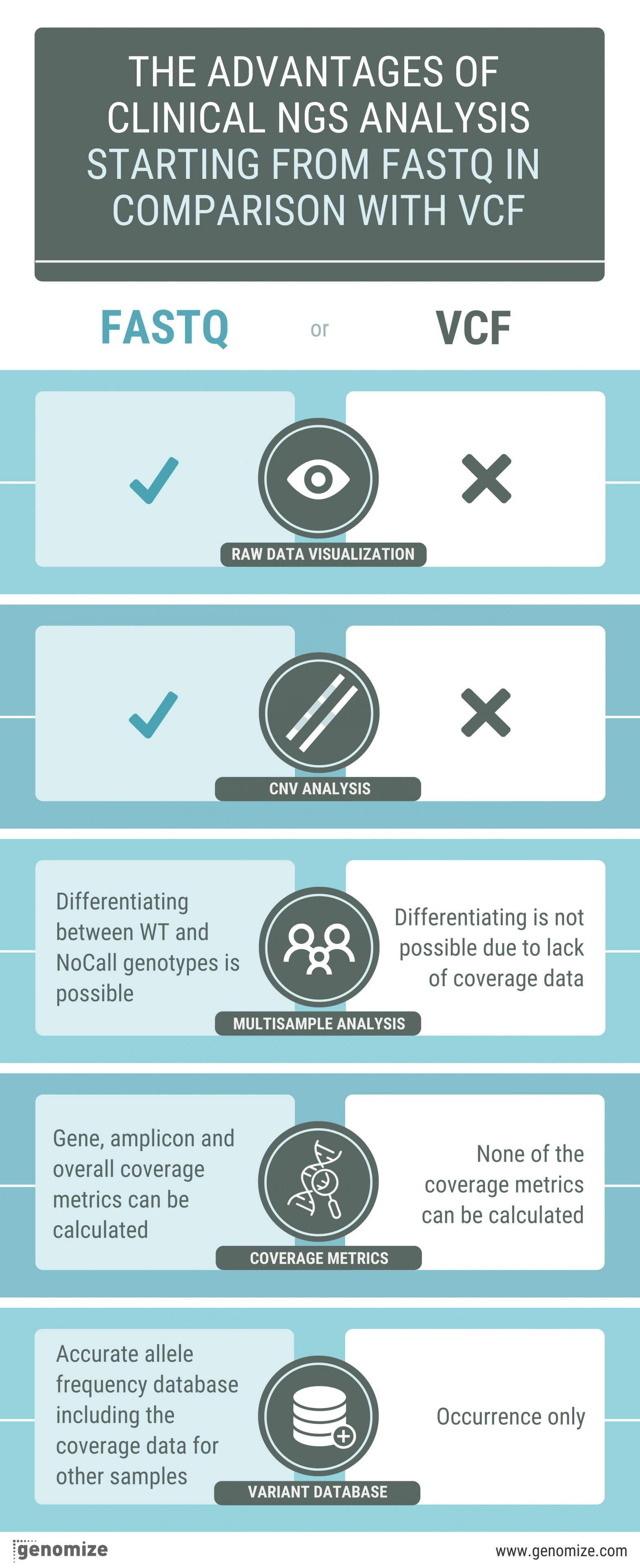
Starting with FastQ files in clinical NGS data analysis has several advantages:
- Raw data visualization: When a clinical analysis is started from the FastQ file, clinicians and researchers can visualize the raw sequencing data, which can provide valuable insights into data quality and potential issues. This is particularly important in a clinical context where data quality and accuracy are crucial.
- CNV Analysis: FastQ files allow for the detection and analysis of copy number variations (CNVs) and other structural variants. This is important in clinical settings, as some genetic disorders are caused by such variations.
- Multisample (Duo, trio, or cohort) Analysis: With FastQ files, you can differentiate between Wild Type (WT) and NoCall genotypes in multisample analysis, whereas it is not possible with VCF due to lack of coverage data. This can help identify inherited or de novo genetic variants that may be relevant for patient diagnosis and treatment.
- Variant database creation: FastQ files enable the creation of more accurate variant databases, including allele frequency and coverage data for other samples. In contrast, VCF files can only provide variant occurrence information.
- Coverage metrics: FastQ files allow for the calculation of various coverage metrics, such as gene, amplicon, and overall coverage. These metrics are essential for assessing data quality and ensuring that the analysis meets clinical requirements. In contrast, coverage metrics cannot be calculated from VCF files alone.
While VCF files are useful for summarizing and reporting known variants, they lack the raw sequencing data and quality scores present in FastQ files. As a result, starting with VCF files in clinical NGS data analysis may limit the scope of the analysis and the ability to identify novel or rare variants.
In summary, FastQ files are a better starting point than VCF files for clinical NGS data analysis because they contain the complete raw sequencing data, including quality scores. This allows for a more comprehensive and customizable analysis, as well as the identification of novel genetic variants that may be important in a clinical context.