The advent of Next-Generation Sequencing (NGS) has revolutionized genomic research and clinical diagnostics by enabling comprehensive analysis of an individual’s genetic information. However, the vast amount of data generated by NGS poses a significant challenge in pinpointing disease-causing variants that underlie a patient’s clinical symptoms. Here, we present an innovative NGS data analysis pipeline available in the SEQ Platform that employs a variant prioritization pipeline integrated with artificial intelligence (AI) algorithms. By leveraging the power of AI, the SEQ Platform effectively filters and ranks variants to produce a shortlist of potentially pathogenic variants, ultimately aiding in the identification of genetic causes for a patient’s clinical presentation.
Whole exome sequencing diagnostic rate is estimated to be about 36% (Fung et al., 2020). In a clinical environment, this figure can change depending on numerous factors, such as the accessibility of supportive data in commonly utilized databases and the breadth of pertinent literature. In some cases, the causative variant is well-documented and classified, resulting in relatively shorter analysis times. In most cases, however, the analyst needs to apply an array of filters, meticulously sift through databases and tools, and conduct an exhaustive review of the literature to pinpoint potential candidate variants. Given the intensive labor involved, variant analysis emerges as the most time-intensive phase of NGS data analysis (Austin-Tse et al., 2022). Nonetheless, the workflow for manual variant analysis, despite being taxing, follows a fairly consistent process. This lends itself well to automation, which can be executed with a high degree of precision using a rigorous and trustworthy variant prioritization pipeline. The automated approach not only elevates accuracy but also significantly reduces the time taken, thereby making the entire process more efficient.
SEQ Platform addresses these challenges head-on. By employing sophisticated AI algorithms, The SEQ Platform streamlines the variant prioritization pipeline, rapidly producing a shortlist of clinically relevant genetic variants that may explain the patient’s clinical symptoms.
We created a 5-tier variant prioritization classification which uses the patient’s clinical information such as the suspected condition, observed phenotypes, expected MOI, age of onset, and sex submitted by the analyst as well as data collected from more than 120 data sources, classifies and ranks each variant based on its potential to be the causative variant(s) (Table 1).
Table 1: Definitions of tiers used in variant prioritization in The SEQ Platform.
We performed a case study to measure the outcomes and benefits of our variant prioritization pipeline using real-world patient data. Whole exome sequencing (WES) samples of 201 patients who visited the clinic* between 2021 and 2023 were included in this analysis (n=201). Their NGS data were previously analyzed using The SEQ Platform but without the variant prioritization functionality. Additional tests and analyses were also conducted as per laboratory and clinic guidelines.
The real-world data set used in this study includes patients whose diagnoses span 102 different diseases. These diseases include but are not limited to, inborn errors of metabolism, various neurological diseases, and various developmental disorders.
For each WES data, the patient’s clinical data is entered into the system, and variant prioritization pipeline was initiated. Samples are then analyzed for distribution of variants within the 5-tier classification (Figure 1) and classification tiers of variants reported by the clinic as causative (Figure 2).
Tiers except for IV and V are designed to include all possibly relevant variants either directly explaining the suspected disease (indicated by the user or automatically deducted from the patient’s phenotypes) or variants that are candidates for other diseases. Our analysis shows 99.89% of the detected variants in a WES sample are categorized as IV or V (Figure 1). This significant number of low-priority variants immediately alleviates the analysis burden and allows the analyst to focus on a very small subset of variants.
Analysis of the variants reported by the clinic as causative showed 97% of the reported variants are classified within the I-III A-B tiers with the majority being included in the IIA classification (Figure 2). ACMG classifications were VUS, VUS+, or VUS++ for 64% of reported variants in tier I-III A-B and 75% for tier IIA variants.
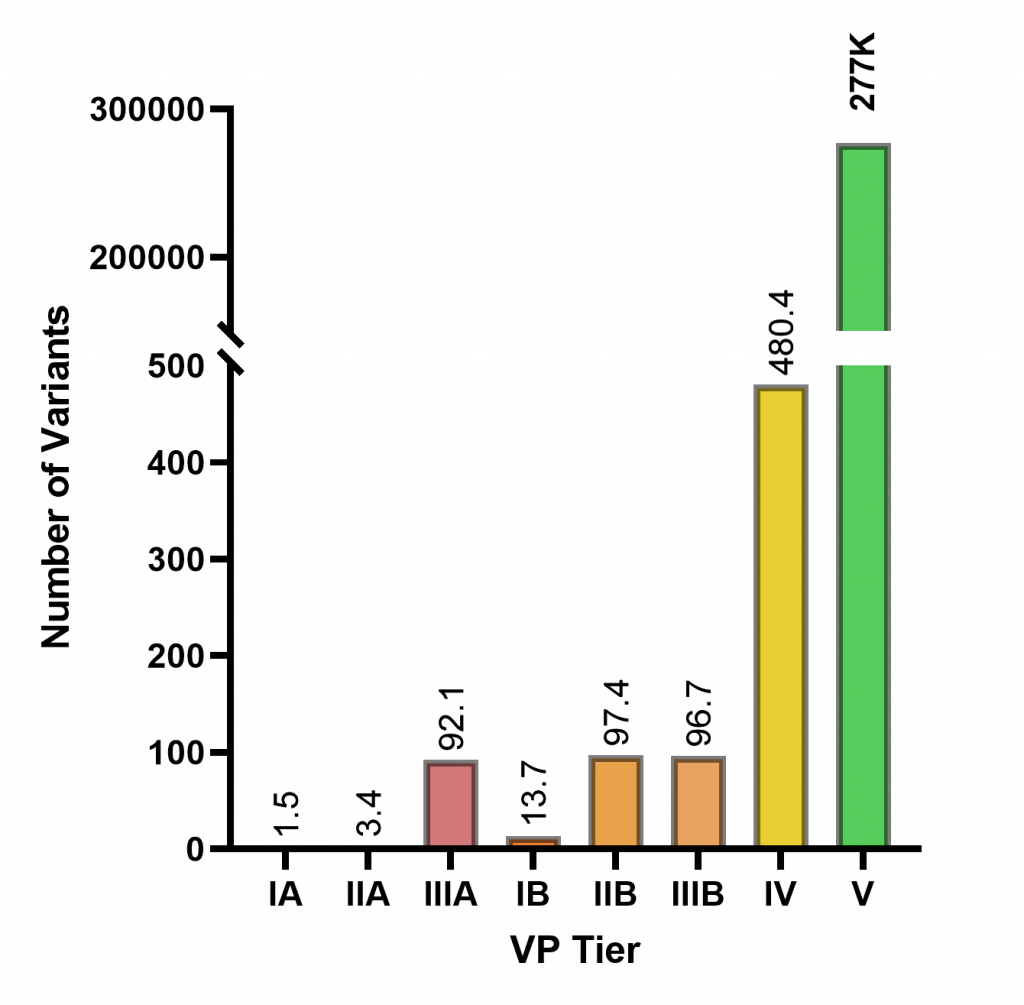
Figure 1: Average number of variants in each variant prioritization tier in a WES data (n=201).
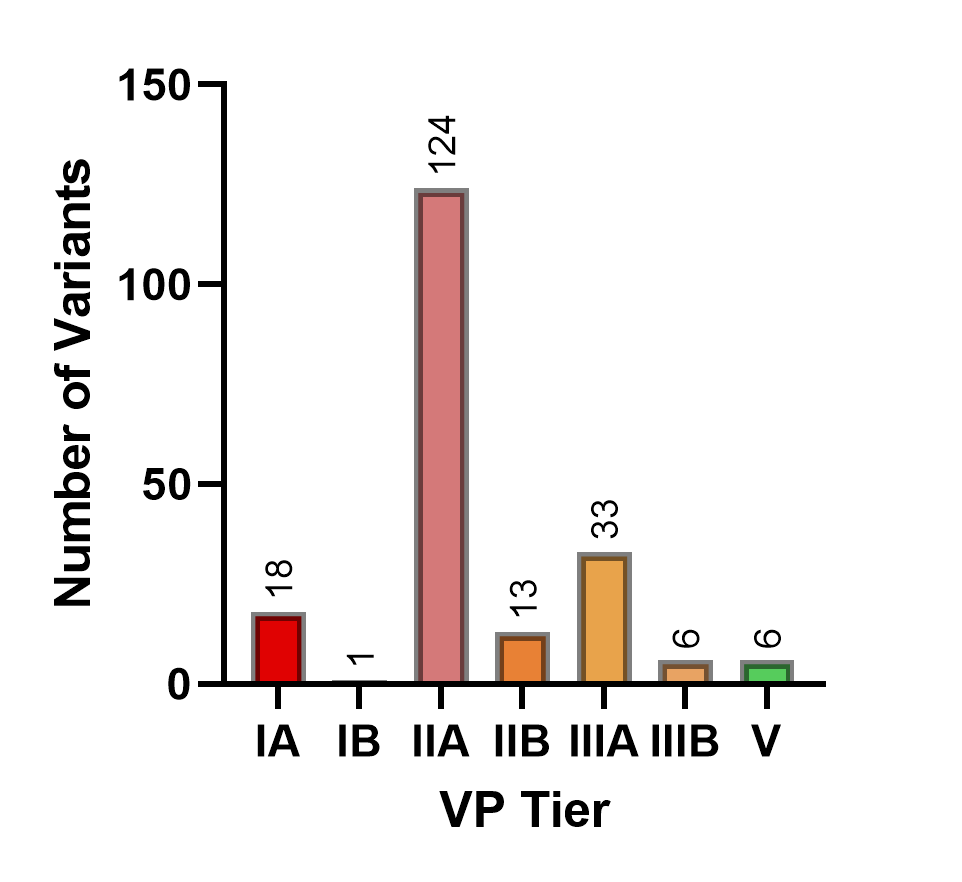
Figure 2: Number of reported causative variants according to the variant prioritization tiers (n=201).
Our aim with The SEQ Platform’s proprietary variant prioritization AI was to significantly reduce the analysis time by combining the clinical information of the patient and the data obtained from NGS, and provide our users with a fast, reliable, and highly accurate variant prioritization.
In our real-world data study, we observed that most of the cases can be resolved within seconds, with the majority of remaining cases only taking a few minutes. A very small minority of cases involve hard-to-diagnose conditions which requires more involvement by the analyst. The comprehensive data we present to our users, however, still greatly improves the analysis times for these cases as well.
In conclusion, The SEQ Platform’s AI-assisted variant prioritization algorithm improves the accuracy of diagnosis, minimizes the likelihood of overlooking crucial genetic variants, and supports clinicians in making more informed treatment decisions.
References
Austin-Tse, C. A. et al. NPJ Genomic Med. 7, 27 (2022). https://doi.org/10.1038/s41525-022-00295-z
Clark, M. M. et al. NPJ genomic Med. 3, 16 (2018). https://doi.org/10.1038/s41525-018-0053-8
Fung, J. L. F. et al. NPJ Genomic Med.5, 37 (2020). https://doi.org/10.1038/s41525-020-00144-x
Wright, C. F. et al. Lancet 385, 1305–1314 (2015). https://doi.org/10.1016/S0140-6736(14)61705-0
Yang, Y. et al. Jama 312, 1870–1879 (2014). https://doi.org/10.1001/jama.2014.14601