With the release 6.0, SEQ Platform now auto-classifies variants detected in a sample, based on ACMG’s Standards and Guidelines for the Interpretation of Sequence Variants.
In 2015, American Collage of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published a joint consensus recommendation for interpretation of sequence variants [1]. ACMG’s recommended standards classify sequence variants, based on 28 criteria, into 5 categories: pathogenic, likely pathogenic, uncertain significance, likely benign and benign. With the new release, SEQ implemented “AutoPathogenicity” feature that classifies variants detected in a sample based on ACMG’s recommended standards, with addition of VUS+ and VUS++ classes. The classification process happens during sample analysis and doesn’t require additional analysis (Figure 1).
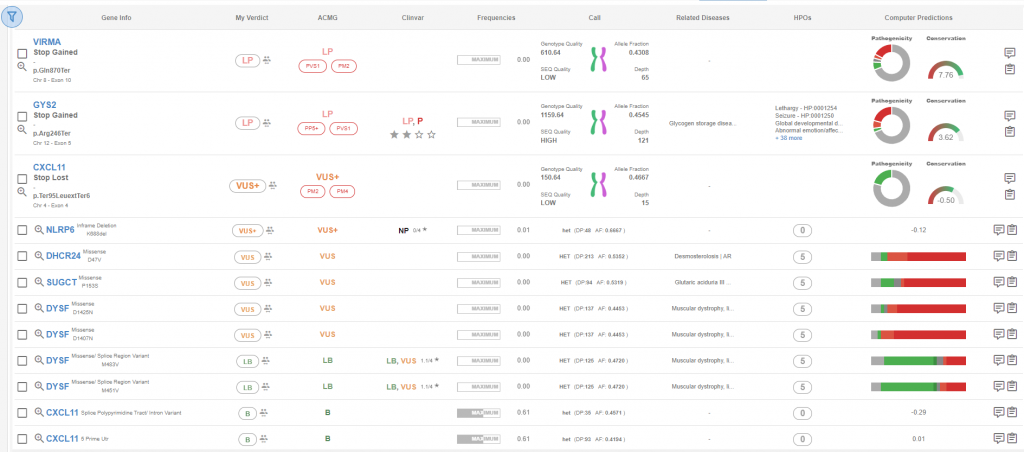
Figure 1: Analysis detail page, sorted by pathogenicity classification.
More detailed information on pathogenicity of the variant can be found in variant details page. In variant details page, under pathogenicity box, given evidence codes (28 criteria that may be used to evaluate pathogenicity of the variants) for the variant can be found as well as unmet and unimplemented codes. The pathogenicity code can be re-calculated by either editing met codes (Figure 2), or adding others (Figure 3).
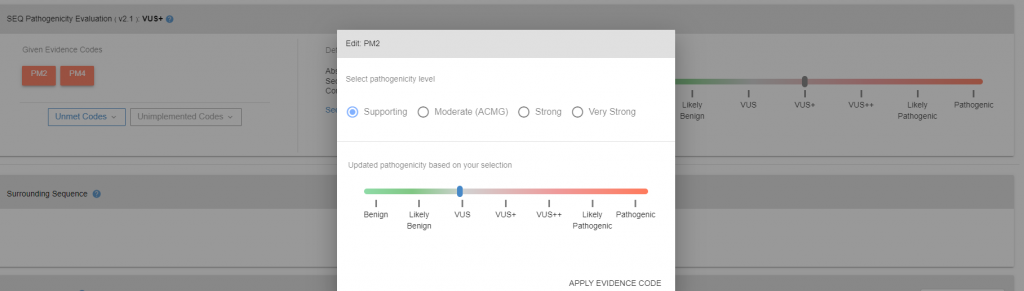
Figure 2: Variant detail page.

Figure 3: Unimplemented evidence codes panel.
VUS+ and VUS++ classes were also added to recommended classification classes to let user see how close a variant to being pathogenic. This is an effort for our users to interpret VUS variants better. We believe VUS+ and VUS++ variants can be prioritized in the case of apparent phenotype or family segregation data.
We appreciate your feedback about our new feature.